After my last post on alternative biochemistries, I thought I’d link to something at least tangentially related, and talk a little about something that’s almost the reverse idea: trying to use the existing DNA information system in our cells to replace silicon-based electronics in computing.
Month: October 2016
Alternative biochemistries…on Earth
Firstly, sincere apologies for the reduced frequency of posting. New job, and new term in full swing (not to mention a lot of baby illness), has meant very little spare time. Things will be slow for a while, but I hope to pick up again more in November.
Lifeforms based on biochemistries other than that found on Earth are a small but firm favourite in science fiction, which is interesting given that so many aliens found in science fiction fall into the “Rubber-Forehead alien” trope beloved of Star Trek. Some of these exotic organisms are just microorganisms, which makes it a bit easier – like the infectious agent in Wyndham’s The Andromeda Strain. I would love to create a truly alien world with fully-fledged thinking aliens based on a unique biochemistry…but I have to say to do so properly would require an enormous amount of research and work (which is to say it would take far more time than I feel I will ever possess!)
The classic is probably the “silicon based lifeform”, perhaps most recognisably in the Horta, the, er, rock beast thing that Spock communed with in Star Trek (after they’d finished zapping it, anyway)

The X-files went one further and had a silicon-based fungus that sent you slightly bonkers (of course). Our life is, famously, “carbon-based” – this is actually referring to the vast majority of organic compounds that are built around carbon and its remarkable talent for bonding with other elements. Silicon, a similar element, is often argued as an alternative, but there are problems with this: silicon is a much larger atom, and it doesn’t form bonds with other elements nearly so readily as carbon does. Moreover, carbon is far more abundant in the galaxy. In fact, on our planet, silicon is the more abundant element, but life arose from carbon anyway.
As an interesting aside, a thermophilic bacterium that lives in hot springs has been found to have a fundamental metabolic enzyme, cytochrome c, that can incorporate silicon into organic molecules, really as an accidental byproduct. Researchers have recently artificially selected this enzyme so that is several thousand times more efficient at this process; not that useful, at the moment, but very interesting nonetheless (and yes, they thought of the Horta too!).
Now researchers are trying to make an organism with a modified genetic code. The genetic code is how the information in DNA is converted into a protein. Three nucleotide bases of a DNA (that sequence of A,T,G and C you’ve all seen) codes (via a “messenger” RNA intermediate) for one amino acid, the building block of a protein. A few encode “start” and “stop” signals for the synthetic machinery. There are about 20 commonly used amino acids in living organims (on this planet), although the code allows for more – there are 64 of these “codons” in fact, and some of them are redundant; that is, they can code for more than one amino acid (you will notice from the table below that it is the last base in the triplet that tends to vary). This reduces the possibility of a mutation in the DNA sequence actually causing a potentially damaging change in the protein.
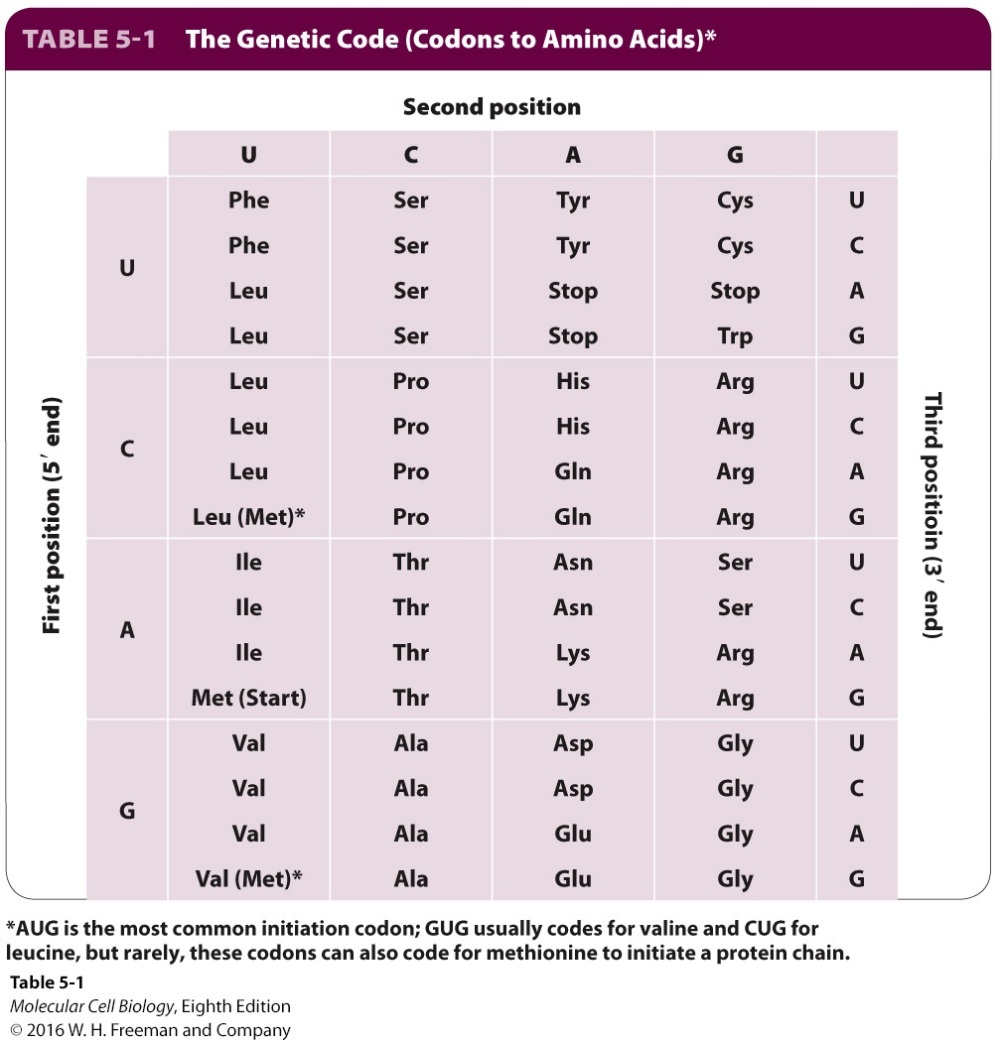
What these researchers have now done is eliminate 7 of the redundant codons in the bacterium E.coli, to leave 57, reasoning that since they were redundant this was unlikely to do the cell any harm. This may sound a little underwhelming, but it is no mean technical feat: they would have to remove every instance of these codons (all 62,214 of them) in the nearly 4 million bases of DNA in the bacterium. Removing them piecemeal would have taken too long, so they essentially re-sequenced the entire genome from scratch.

Figure 2A: Codons AGA, AGG, AGC, AGU, UUA, UUG, and UAG were computationally replaced by synonymous alternatives (center). Other codons (e.g.,UGC) remain unchanged. Color-coded histograms represent the abundance of the seven forbidden codons in each segment.
Why bother? Like a lot of speculative science, it’s a little hard to tell how useful it will be, but there are a lot of potential uses. E.coli is used to synthesise a lot of proteins useful to us, like a little bioreactor, and this could render it immune to infection by viruses that depend on the codons it no longer uses. Additionally, these seven removed codons could now be used to code for a new synthetic amino acid not normally found in nature, potentially opening up a world of novel proteins. (Oh, and yes, they did build in a failsafe). This is still a work in progress; the genome hasn’t been completely assembled yet, but it’s an interesting and conceptually radical idea. It’s not a non-carbon based biochemistry, to be sure, but, if it were taken a few steps further, it could mean we could create an entirely synthetic organism with an entirely different genetic code to our own. And that’s pretty science fiction, if you ask me.
Reference
Ostrov et al, 2016: Design, synthesis and testing toward a 57-codon genome, Science Vol. 353, Issue 6301, pp. 819-822 DOI: 10.1126/science.aaf3639
